Quantitative Cell Biology
Understanding the Dynamics & Functionality of a Population from Singlce-cell Behaviours Learn MoreQuestions & Approaches
Simple single-cellular organisms such as Escherichia coli, yeast, and cultured cells are good model systems to elucidate the quantitative laws and principles of biological systems. We are collaborating with various experimental biologists to unveil such principles by integrating different types of quantitative data with mathematical modelling and informatics approaches. In particular, we are focusing on how population-level behaviours emerges from the combination of single cell-level behaviours of cells and their heterogeneity.
01.
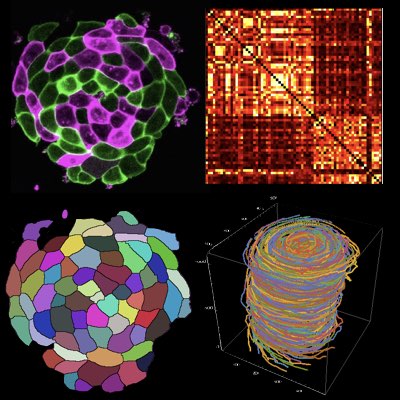
Single-cell Dynamics of Epigenetic Memory
Epigenetics is a biological mechanism by which information is transmitted from one generation to another without change in the genetic sequence, which is believed to increase the adaptability of the cells in a slowly changing environment. To understand how epigenetic memory is maintained and transferred to descendent cells, we have monitored and traced the epigenetic states of yeast cells at the single-cell level over several generations. By using a simple statistical method, we have estimated the stability of the epigenetic memory and verified the correlation between epigenetic states on different regions on the genome (Learn more)
02.
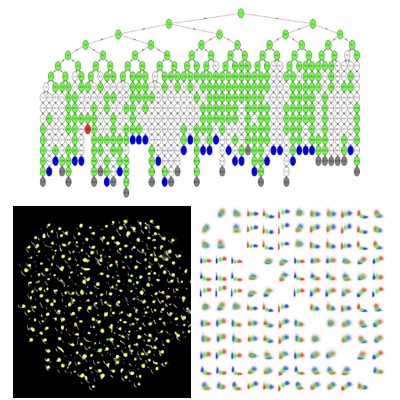
Singularity & Desynchrony of the Circadian Rhythm
The internal timing of multicellular organisms is shaped by the collections of single cell-level cellular clocks. However, at the population level, the response of the clocks may not be simply determined by the intrinsic behaviours of individual clocks. One pivotal example is the singularity of the circadian rhythm, in which the rhythm is stopped by an external perturbation applied at a specific time point with appropriate strength. This rhythm cessation may be attributed either to stoping of the clock in individual cells or to the desynchrony of the cells. By combining a high-throughput squeening and single-cell imaging systems, we have demonstrated that perturbation-induced desynchrony is the primary mechanism of the singularity. Moreover, using a mathematical model, we demonstrated that the population-level response of circadian clocks can be explained quantitatively by combining a single cell-level response and heterogeneity of the population(Learn more: [1], [2]).
03.
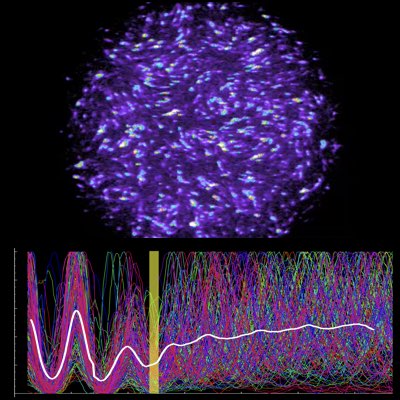
Linking Single-cell Chemotaxis with Collective Migration
Cell-cell interaction is a mechanism that enables multi-cellular organisms to coordinate the function and behaviour of their cells as a single unit. To understand how cell-cell interactions shape and control the complicated behaviours of cells, we studied chemotaxic amoeba cells, which communicate via cell-cell and chemical interactions. By using an information-theoretic technique, we have identified two subgroups of cells that have different kinetic properties from the tracking trajectories of cells in the population. This approach may also be used for analysing the more complicated patterns of cells in organisms.
04.
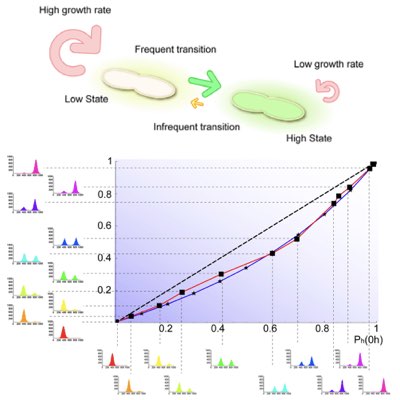
Dissecting the Impacts of Stochastic Dynamics & Growth
Single-cell behaviours are commonly inferred by using population-level measurements, which are relatively more simple than single-cell observations. However, the single-cell behaviour may be qualitatively different from the population-level behaviour, especially when cells show cell-to-cell variability and when different phenotypic states are subject to selection. We demonstrated how the interplay between such intercellular heterogeneity and selection can induce a huge discrepancy at the single-cell and population level by using synthetic toggle switches as a model system (Learn more).