Questions & Approaches
Establishment of a mammalian pre-implantation embryo is the first developmental process during which a single fertilized egg differentiates into different types of cells, including pluripotent cells, which generate all of the cell types in our body through subsequent developmental processes. To understand the mechanism by which the complex multicellular structure of the body is formed spatially and dynamically, it is essential to be able to track and reconstruct developmental lineages. To this end, along with our collaborators, we are developing a long-term and quantitative time-lapse 3D imaging system, automatic cell detection and tracking algorithms, statistical methods to categorize stochastic developmental lineages, and models of embryogenesis. Our methodologies will further contributes to quantifying the states of embryos in reproductive assistance technologies so as to best optimize culture conditions, which critically impacts the success of the artificial insemination (Learn more).
01.
Automatic Detection of Nuclei from 3D Imaging Data
The first step for the lineage reconstruction of a developing embryo is to detect the positions of cells from 3D imaging data. Starting from simple algorithms combining basic and traditional image processing filters, we have developed more complicated algorithms based on integer programming and deep neural networks by feeding grand-truth data obtained from the basic algorithms into more sophisticated algorithms as training data sets (Learn more: [1], [2]).

02.
Cell Detection by Deep Learning
In bioimage analysis, we typically have to reset and adjust control parameters when the quality of the data or the conditions of measurement is changed. Although it can be automated to a certain extent, such process is difficult and requires experience especially if we work on 3D images. We have developed a new deep learning-based method for identifying cell nuclei from 3D images. This makes it possible to analyze a variety of embryos from different species acquired under various conditions. (Learn More)。
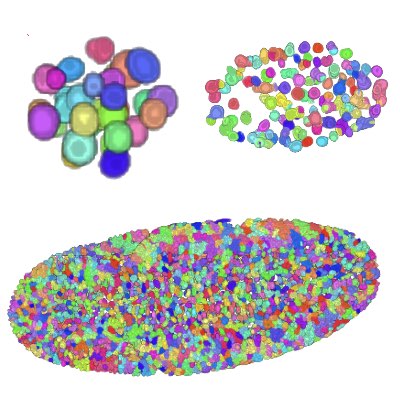
03.
Tracking Dividing Blastomeres
The tracking of dividng cells becomes exponentially more difficult as the number of cells increases through cell divisions. In achieve robust tracking of dividing cells, we have formulated tracking as an integer programming to obtain a very accurate algorithm. The score of the association of cells is then further improved by employing machine-learning techniques.

04.
Categorizing Developmental Lineages
For comparison of the developmental processes of embryos, the task can be reduced to the problem of comparing lineage trees corresponding to the developmental processes of individual embryos. However, statistical and machine-learning methods for such data are still lacking, mainly because temporally growing branching systems are not common outside of the field of biology. Using the path-integral formulation of population dynamics, we are also constructing new statistical methods to extract the quantitative properties of dividing cells from lineage data.
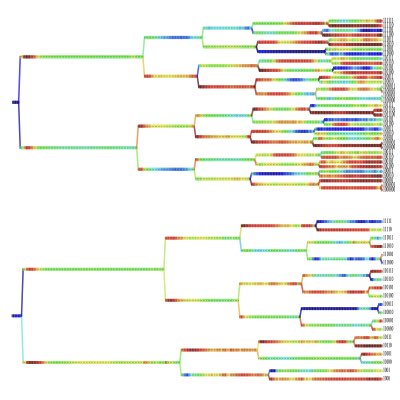
04.
Modelling Embryogenesis
The spatial positioning of cells in an embryo is an important determinant of the ultimate cell fate. To extract how the positioning and fate of cells are jointly determined through the combination of gene expression and mechanics, we are developing a mathematical model of embryogenesis, which can be integrated with the lineage data obtained by tracking the imaging data.