Our Projects
Goal, Approaches, & Biological TargetsUnderstanding Biological Information Processing
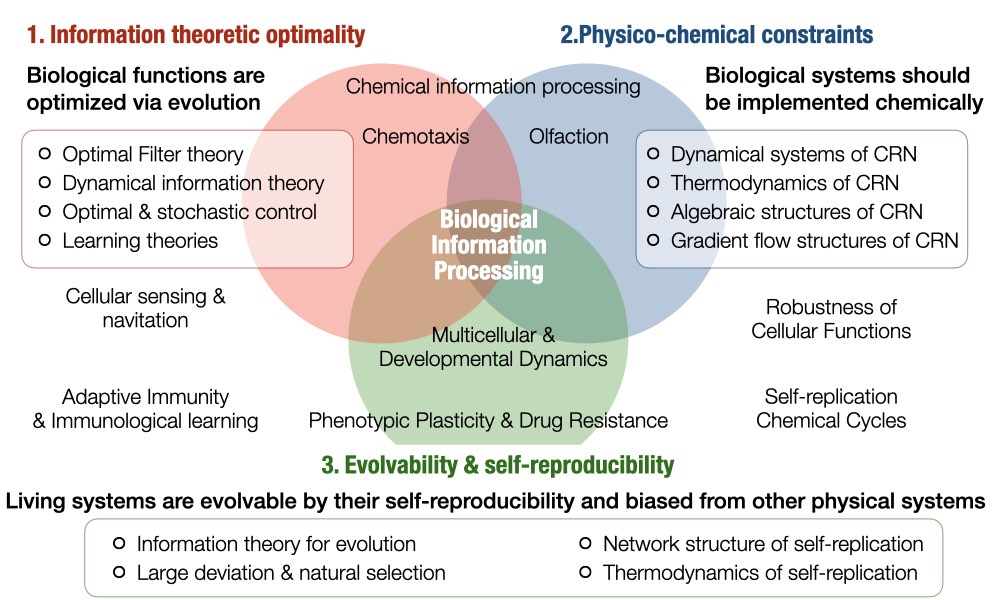
In order to understand biological information processing systems created by evolution, we focus on their (1) information theoretic optimality, (2) physico-chemical constraints, and (3) evolvability & self-reproducibility. To grasp such aspects, we are working on
- Construction of relevant mathematical theories (Learn More)
- Development of informatic methods (Learn More)
- Analysis of various biological phenomena (Learn More)
Mathematics
In order to describe and understand information processing in living organisms, mathematical methods and theories that can capture the essence of the phenomena are necessary. While many theories and techniques have already been developed in the fields of mathematical science, physics, and engineering, they are rarely applicable to biological phenomena as they are. We need to modify, improve, and in some cases, recreate them for our purpose. Specifically, we are using and developing the following theories in our works:
Dynamical Systems(DS)
- Deterministic & stochastic DS
- Deterministic bifurcation theory
- Stochastic bifurcation theory
- Gradient flow
Stochastic Processes
- Point processes
- Diffusion processes
- Conditoned stochastic processes
- Large deviation theory
Information Theory
- Dynamic information theory
- Optimal filtering theory
- Sequential decision theory
- Information geometry
Thermodynamics
- Chemical thermodynamics
- Stochastic thermodynamics
- Information thermodynamics
- Thermodynamics of evolution
Control & Learning Theories
- Optimal control theory
- Stochastic control theory
- Statistical learning theory
- Reinforcement learning
Algebra & Geometry
- Algebraic graph thoery
- Algebraic homology
- Hessian geometry
- Algebraic geometry
Informatics
Quantitative measurement techniques for biological phenomena have made great strides in the past two decades, providing us with the data we need to investigate phenomena in detail and to test theories. Such new and unique data provides us with the venue for developing new informatic methods and theories. We are also developing a variety of informatics methods to extract appropriate information from these quantitative data and integrate them with theories:
Statistical Analysis
- Inference & prediction of population dynamics equation
- Inference & prediction of histry-dependent point processes
- Inference & prediction of hidden states of cells
Numerical Simulations
- Simulation of deterministic & stochastic equations
- Simulation of multi-cellular developmental processes
- Simulation of immunological learning processes
Discrete Data Analysis
- Theory of flow & dynamics on graphs
- Extensions of graph NN
- Inference of information flow on graph
Bioinformatics
- Machine learning of T-cell receptor repertoires
- Machine learning of Raman-Omics correspondance
- Machine learning of Multi-Omics correspondancee
Chemical Information Processing
- Representation and ML of odor molecules
- Machine learning of olfactory receptor responses
- Design and prediction of flavor of odor mixtures
Analysis of Biological Phnomena
A close examination of biological systems let us find that even seemingly simple single cells possess breath-takingly sophisticated and efficient functions. Toward understand information processing in cells, we are working on the mathematical understanding of various phenomena in collaboration with our collaborators:
Chemotaxis
How do cells sense dynamic environmental changes from stochastic sensory-receptive signals and select appropriate behaviors from noisy sensory information?
Chemotaxis is the simplest and ideal system to address these questions.
We are addressing the above questions by combining theoretical modeling, information theory, and optimal control theory with quantitative data of
Adaptation & Phenotypic Plasticity
Cells actively adapt to changing environments by changing their phenotype. At the same time, cells happen to be adaptive in a population are passively selected. We are investigating how these two types of adaptation and selection lead to the evolution of cells and cell populations, using genealogical tracing in systems such as E. coli and phenotyping using Raman imaing in combination with theoretical models (Learn More).
Chemical Sensing & Olfactory System
The olfactory system processes combinatorial chemical information consisting of a complex mixture of odorants. How is chemical information received, recognized, and processed? We address these problems by combining theory and quantitative data science using receptor response and sensory measurement. We are also conducting joint research with companies for application.
Adaptive Immunity & Immuno-repertoire
Adaptive immune system remembers past infections and responds appropriately in subsequent infections. How does the immune system recognize unknown foreign enemies and realize learning of appropriate immune responses from past infections? We are working on prediction of our infection history and immune status by decoding sequence data of immune receptors?(Learn More)。
Developmental Lineage and Dynamics of Embryo
Development is the process by which diverse cell groups and spatial structures arise from a single cell. What are the mechanisms and principles behind it? In addition, how can we reconstruct developmental dynamics and genealogical information from quantitative image data? We are tackling these questions by building new methods and models of deep image analysis, genealogical analysis, and developmental models(Learn More).
Functions and Control of Collective Phenomena
Cells and organisms operate both as individuals and as groups to achieve complex and functional behaviors. How do groups of cells mutually regulate each other's behaviours and realize vairous functions such as group exploration? Also, how do organisms resolve the disagreement of interests that arises between individuals and groups? We are interested in and working on these questions (Learn More).
Self-replication
Self-replication is a unique property of life and a prerequisite for evolution. The necessary and sufficient conditions for self-replication to occur in chemical systems are still unknown. We are also investigating the problem of realizing self-replication with chemical reactions and the properties that self-replicating systems universally retain as a result(Learn More).
Signal Processing by Cells
It has been suggested that the specificity of individual reactions in cellular reaction networks is not as high as previously thought, and that there is confounding and interference in information transmission and metabolic catalysis. We are also addressing questions such as how do cells transmit and process diverse information in a reaction system where confusion and interference are inevitable?